B5: FIND - An open-source analysis toolbox for ensemble spike recordings
Karl-Heinz BovenE, Ad AertsenA and Ulrich EgertP
E = Multi Channel Systems GmbH
A = Neurobiology and Biophysics
P = Biomicrotechnology
Scientific background
The idea of dynamically organized cell assemblies as building blocks of computational processes underlies most current thinking about higher brain function. Thus, recent years show a growing interest in simultaneously recording spike trains from multiple (now up to a few 100) neurons. With this technical progress, there is growing need for new computational tools to analyze and interpret the large data flow from these experiments. Indeed, progressing from single-neuron to network physiology implies a non-linear increase in complexity of data analysis. Appropriate concepts and methods to assess cooperative dynamics in large populations of spiking neurons are still lacking and need urgently be developed. While there is undeniable progress in novel analysis methods implementations remain scattered in the literature or hidden in (ill-documented) software collections. A unified, well-documented interface to such tools would greatly enhance the efficacy of experiments and improve possibilities to compare results across experiments and labs.
Objectives
We will develop a platform-independent toolbox to analyze spike data from single- and multiple-electrode recordings in vivo by providing a set of standard and more advanced analysis methods, building on experience in design and application of such methods and open toolboxes, and excellent working relations with top laboratories in the field. The toolbox will accommodate import of multiple proprietary data formats, based on the Neuroshare Project, www.neuroshare.org. Thus, physiological data from different acquisition systems can be compared using identical analysis methods, and results can be verified across experiments and laboratories. To enable the incorporation of new algorithms - a weakness of most commercial toolboxes - our toolbox will be open, providing the possibility to extend the collection of algorithms with new ones will facilitate the development and distribution of new techniques among the scientific community. Exchange and discussion will be organized by an Internet site, including up- and download areas and forums between developers and users, and by organizing tutorials and training workshops.
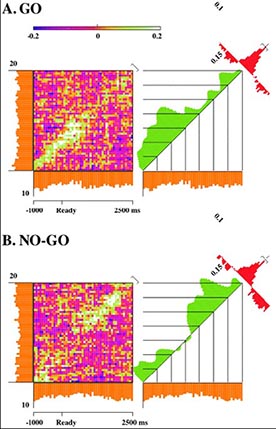
Fig. 1. The Joint-Peri-Stimulus-Time Histogramm (JPSTH) measures the dynamics of spike correlation in a pair of neurons after correcting for rate modulations within trials and rate variability across trials (Aertsen et. al. (1989) JNeurophysiol 61:900-17). Here, JPSTH-analysis was used to reveal dynamic modulation of correlated firing between frontal cortex neurons in relation to behavioral events (From: Vaadia et. al. (1995) Nature 373: 515-518).
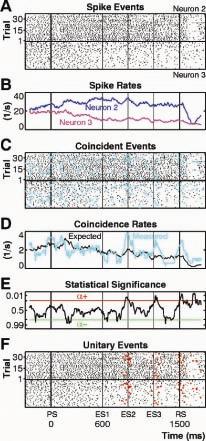
Fig. 2. Unitary Event (UE) analysis detects the occurence of conspicuous patterns of coincident joint spike activity among groups of simultaneously recorded single neurons (Grün et. al. (2002) Neural Computation 14: 121-153). Here, UE-analysis was used to reveal Instances of accurate spike synchronization in monkey primary motor cortex in relation to external events (stimuli, movements) ans to purely internal events (stimulus expectancy). (From: Riehle et. al. (1997)
Science 278: 1950-1953).

